Synthesis, Structure, and Biological Activity of des-Side Chain Analogues of 1 ,25-Dihydroxyvitamin D3 with Substituents at C-18
Verlinden, L., Verstuyf, A., Eelen, G., Bouillon, R., Ordonez-Moran, P., Larriba, M.J., Munoz, A., Rochel, N., Sato, Y., Moras, D., Maestro, M., Seoane, S., Dominguez, F., Eduardo-Canosa, S., Nicoletti, D., Moman, E., Mourino, A.(2011) ChemMedChem 6: 788-793
- PubMed: 21520419
- DOI: https://doi.org/10.1002/cmdc.201100021
- Primary Citation of Related Structures:
3P8X - PubMed Abstract:
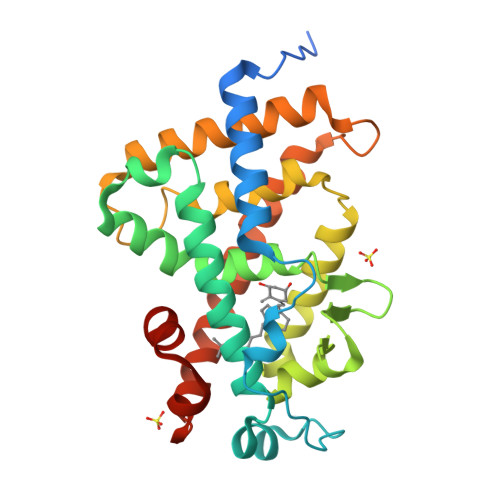
An improved synthetic route to 1α,25-dihydroxyvitamin D(3) des-side chain analogues 2 a and 2 b with substituents at C18 is reported, along with their biological activity. These analogues display significant antiproliferative effects toward MCF-7 breast cancer cells and prodifferentiation activity toward SW480-ADH colon cancer cells; they are also characterized by a greatly decreased calcemic profile. The crystal structure of the human vitamin D receptor (hVDR) complexed to one of these analogues, 20(17→18)-abeo-1α,25-dihydroxy-22-homo-21-norvitamin D(3) (2 a) reveals that the side chain introduced at position C18 adopts the same orientation in the ligand binding pocket as the side chain of 1α,25-dihydroxyvitamin D(3).
Organizational Affiliation:
Laboratorium voor Experimentele Geneeskunde en Endocrinologie, Katholieke Universiteit Leuven, Leuven, Belgium.